Systematics
Comparative analysis among selected SGHV annotated genes has shown that these genes are distinct from homologues present in other dsDNA insect viruses. For example, Garcia-Maruniak et al., (2009) compared SGHV sequences to a series of core genes present in baculoviruses and nimaviruses and found that in the majority of cases the SGHVs clustered as a distinct group with little relatedness to other insect dsDNA viruses.
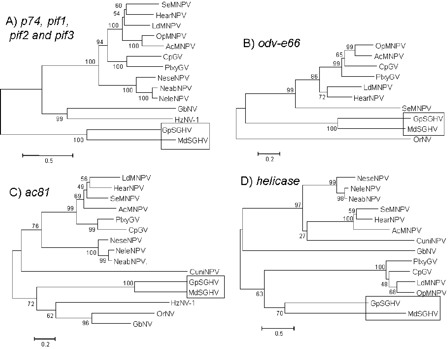
Neighbor-Joining (NJ) phylogenetic trees of (A) combined p74, pif-1, pif-2 and pif-3, (B) odv-e66, (C) ac81, and (D) helicase proteins and their homologs in baculoviruses and nimaviruses. The following viruses were included: Lepidopteran-specific NPVs: A. californica (Ac) MNPV, O. pseudotsugata (Op) MNPV, L. dispar (Ld) MNPV, S. exigua (Se) MNPV, H. armigera (Hear) NPV; Granuloviruses: C. pomonella (Cp) GV, P. xylostella (Plxy) GV; Hymenopteran-specific NPVs: N. sertifer (Nese) NPV, N. lecontei (Nele) NPV, N. abietes (Neab) NPV; Dipteran-specific NPV: C. nigripalpus (Cuni) NPV; and Nudiviruses: H. zea (Hz) NV-1, G. bimaculatus (Gb) NV, O. rhinoceros (Or) NV. Numbers on the nodes indicate bootstrap values (500 replicates). Taken from Garcia-Maruniak et al., (2009). Click on image to enlarge.
More recently, a phylogenetic analysis by Abd-Alla et al. (2009) of DNA polymerase sequences from a wide range of insect dsDNA viruses reiterated the identity of the SGHVs as a unique viral family.
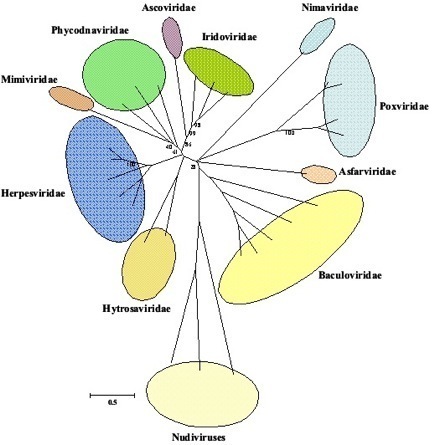
Neighbor-Joining (NJ) trees of DNA polymerase predicted amino acid sequences based on 2374 sites from 29 viruses from 11 families. Ascoviridae: Spodoptera ascovirus; Iridoviridae: Lymphocystis disease virus 1, Invertebrate iridescent virus 6, Aedes taeniorhynchus iridescent virus; Nimaviridae: shrimp white spot syndrome virus; Poxviridae: Amsacta moorei entomopoxvirus, Choristoneura biennis entomopoxvirus, Vaccinia virus, Molluscum contagiosum virus; Asfarviridae: African swine fevervirus; Baculoviridae: Culex nigripalpus NPV, Neodiprion lecontei NPV, Cydia pomonella granulovirus, Autographa californica nucleopolyhedrovirus, Lymantria dispar nucleopolyhedrovirus; Nudiviridae: Heliothis zea virus, Oryctes rhinoceros virus, Gryllus bimaculatus nudivirus; Hytrosaviridae: Glossina pallidipes salivary gland hypertrophy virus, Musca domestica salivary gland hypertrophy virus; Herpesviridae: Human herpesvirus 6B, Porcine cytomegalovirus, Bovine herpesvirus 1, Retroperitoneal fibromatosis-associated herpesvirus, Macaca fuscata rhadinovirus; Mimiviridae: Acanthamoeba polyphaga mimivirus; Phycodnaviridae: Paramecium bursaria Chlorella virus 1, Ectocarpus siliculosus virus 1, Emiliania huxleyi virus 86. Taken from Abd-Alla et al., (2009).
Virus Biology









Systematics




In addition to comparisons made among selected SGHV genes, Garcia-Maruniak et al. (2009) generated syntenic maps of MdSGHV, GpSGHV, and several nudiviruses to compare their general genomic organization and to visualize conserved regions of both coding and non-coding DNA. The syntenic map between MdSGHV and GpSGHV showed significant co-linearity between the two genomes. Syntenic analysis of the MdSGHV and GpSGHV genomes identified a total of 161 conserved, non-ORF regions sharing amino acid sequence identities of between 25% and 73%. When this analysis was extended to include HzNV-1 and GbNV, the number of conserved regions was reduced, and there was a lack of co-linearity of these viruses with the SGHVs. These results strongly suggest that GpSGHV and MdSGHV are related, and are distinct from other large invertebrate DNA viruses.
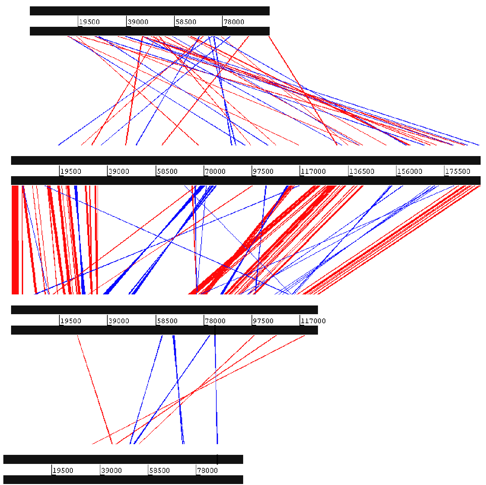



Syntenic maps comparing the overall level of colinearity of the Musca domestica salivary gland hypertrophy virus (MdSGHV) and Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) genomes. Blue lines indicate inversions, and red lines indicate identity among high-scoring pairs. Bands are between conserved regions, not necessarily ORFs. Genome position in bp is shown between the lines of each dsDNA viral genome. The number of conserved regions does not reflect the level of homology in each region. Note the low identity between the SGHVs and nudivirus.